Updated 28-Jun-2024
This is part of McNeill and Beyond - a Memoir
If we go back far enough, our ancestors are from Africa, as are everyone else's. For Y-DNA (our patrilineal line) this means ~ 110,000 years ago there is what we would call an anthropological commonality. Between the anthropological and the genealogical (stories of families) there is the historical, which ranges from hundreds to tens of thousands of years, basically that of archaeology.
If we are to talk about a meaningful genealogical timescale, something on the order of human history is needed, not archaeology or anthropology. The story of families is one entwined with that of the story of homo sapiens and individual people. Therefore, oral and written history is needed as a backdrop.
However, it is true that archaeological/historical time horizon remains are many times considered a part of human meaning and have value. Essentially this corresponds roughly with the bronze age, beginning around 6,000 years ago, but of course recedes into the more distant past with any such evidence of human habitation.
Note: Y-DNA and MT-DNA analysis is very different from Family Finder and other similar tests which aim to show percentages of a given ethnicity for an individual. Findings on twins have been shockingly inaccurate. There is a term for this, ancestry tests of this kind have been labelled genetic astrology. Even findings within the past year (2019) are still flawed (largely because of a lack of data).
What we also mean by lands of our ancestors is the journey from one place to another. Those journeys and their impetus can be as interesting as the years and generations in a single place. And so we need a way to trace our journeys and our respites in various places, but the story-telling can now be supplemented with the tales written by our very bodies, our genetic code.
And so we will deal with three kinds of history: family history, that is the stories of descent. This is the timescale of several hundred years. Then next will be a clan and national histories, in which last names are present. This goes back a few hundred years or so, though there are claims to greater antiquity. Third is genetic genealogy which is in the timescale of thousands to tends of thousands of years, and is based on chromosome mutation rates.
History and Genetic Genealogy
Genetic Genealogy is the use of genetic mutation rates to determine divergence and similarity, and so can help trace the relationships between people, and groups of people, over a fairly long time-scale that has been lost in other ways. We can find out how related we are to others over hundreds and even thousands of generations, and therefore the journeys which our ancestors took, even starting back in Africa.
Genetic Genealogy is the study of families using analysis of DNA. There are two kinds of DNA to study this, Y-DNA is part of the Y chromosome (passed from fathers to sons), and MTDNA which is passed on from Mothers to their children. Both males and females have X chromosomes (females have two, one from each parent), whereas males have an X and a Y, which is why good genetic tests are done with DNA from males. For females who want testing of the Y-DNA, it is most accurately done with DNA from a male sibling who share both parents.
Genetic Genealogy deals with hundreds to thousands of years, whereas genealogy alone (the stories of families) best deals with tens to hundreds of years (10 generations or so). One thing about genealogy is that stories can be wrong (and so-called non-paternity events can take place, where the actual father of a child is not known or hidden).
Genetic Genealogy can't lie, as long as samples are taken accurately and not contaminated. However, it is less accurate in terms of dates and places as we are dealing with a longer time scale and the variability of mutation.
It turns out that both for Y-DNA -- inherited from father to son -- and MTDNA -- inherited from mothers -- there are less than 20 root ancestors. These are coded by letters. Indeed, there is a Y-chromosomal Adam (as well as a Mitochondrial Eve), a common ancestor to all modern humans, whose chromosomes are from 200,000-300,000 years old (for Eve it is 99,000-200,000 years).
Note that this does not mean everyone is descended from this Y-chromosomal Adam, but descent from other humans are not solely through the Y-chromosome. The implication is that this Adam had at least two sons who both have unbroken lineages to the present day. This is the definition of most recent common ancestor Y-MRCA, that is two individuals have an ancestor in common (namely, Adam).
Genetic genealogy is very different from regular genealogy, with the family tree and multitude of branches. For one thing, with Y-DNA analysis, only the patrilineal line is available (father's father's father's father's father, etc.) which means it really only deals with a direct line (a single branch) in the family tree.
Secondly, there are basically three kinds of analysis going on in the standard packages and tests:
- Y-DNA - Patrilineal only
- Haplogroup (SNP) - most recent regional marker (generally a few thousand years ago, e.g., Angles and Saxons)
- STR genetic marker matching between two samples (I see we are matched on 67 markers with a genetic distance of 2), calculates sharing of most recent (male) common ancestor generation probability.
- MTDNA - Matrilineal only
- Autosomal DNA (inherited from both parents) this says what percentage is from what part of the world)
The Autosomal DNA tests are now the most common and the results say such things as 10% East Indian, 20% African and the like. This seems to be misleading, as it is interpretable only in terms of current, modern racial/nationalist sentiments (while not overtly racist, it requires these categories for meaning). In any case, all the genetic research here is based solely on Y-DNA testing.
Genetic Bottlenecks, Culture, and Writing
The human species may be 200,000 years old or so (though alternative approaches make claims of up to 1 million years). Full human modern behavior (culture, from the anthropological perspective) appeared around 50,000 years ago. The surviving species emerged from Africa after the Toba Eruption Event around 69-77,000 years ago.
Toba is suggested as a near-extinction event, which could have killed off any other humans who may have been alive before then, though there is now evidence that some local populations in equatorial areas did survive (Flores Man).
Whatever the cause (Toba event or not) there is a known hourglass or bottleneck where the human species was likely reduced to around 10,000 breeding pairs (some say 7,000).
Africa may not have been the original place of origin of the species, but is generally thought to be the starting point after Toba/hourglass. Most Haplogroup time estimates are shorter than 50,000 years, and so here we enter the genetic genealogical time frame. Humans took up different routes at different times out of Africa, and this kind of haplogroup wandering can be found in the fossil record.
Some time around 10,000-12,000 years ago some human groups left the hunting and gathering lifestyle and innovated with wild grains but turned into true agriculture (and animal husbandry, which may have occurred before then).
Around 6,000 years ago the Bronze Age /Late Neolithic ushered in advanced mining, smelting and metalworking. Evidence of Bronze technology continued as late as 500 CE, such as in Thailand.
Around 5,000 years ago writing technology also first appeared (in Sumeria). In places like Ireland, scripts such as Ogham could be a little over 2,000 years old, about the same time that the 500 year old Buddhist canon was being committed to writing in Pali in Sri Lanka, South Asia.
In terms of Southeast Asia, the written Cham language in Vietnam appeared around 1,800 years ago. Khmer had a script (based on Pallavi) around 1,500 years ago. Myanmar, Thai and Vietnamese writing is of more recent origins, around 700-800 years ago (possibly a little longer).
Six hundred years ago, the moveable type printing press came into existence, having an enormous impact on literacy, and hence human culture and behavior (though it took hundreds more years to produce mass literate education).
Human history and culture flows from the bottleneck of 70,000 years ago, to animal husbandry and agriculture, to metalwork, to writing technology. Our genetic history changed throughout this time period with internal mutation rates. Our people migrated to different places, living and moving along. At first they may have followed animal migration patterns, or were forced to move based on changing resources, and the threat of attack from larger groups.
Note for much more extensive history that stretches far beyond hominid, and even mammalian history, see The Ends of the Earth.
The McNeill Haplogroup R1b1a1b1a1a1c2b2a1b5a
(by McNeill I mean this McNeill, not all McNeills)
If we look at the origins for our McNeill Haplogroup, we begin the story around 10,000 years ago when the R1b people are purported to have been the first Neolithic cattle herders. R1b1a2 (M269) were Caucus people and reached central asia. These people genetically are also associated with the diffusion of Indo-European languages. the R1b1a1a2a (L23) mutation is somewhere between 6,000-4,0000 years ago in the Caucuses. However, it is unclear at which point these people traveled eastward.
And so we are at the late neolithic/bronze age, at time of Standing Stones, proto-written languages, and a diversity of people interacting through trade, migration and warfare. Events in history begin to be chronicled in a way that can be verified (e.g., the Trojan War as told by Homer was an actual event approximately 3,000 years ago).
R1b1a1a2a1a1a (S21/U106) occurred somewhere during the Bronze age roughly around 4,000 years ago. This is associated more closely with Scandinavia (essentially the Y-DNA mutation and specific migration patterns occurred coincidentally).
- R (R-M207), 52,000-81,000 years ago1
- R1 (R-M173), Central Asia, 12,000-25,000 years ago
- R1b (R-M343)...
- R-M269 (R1b1a1a2)
- R-U106/S21 (R1b1a1a1a2a1a1), ~5,400-4,500 years ago
- R-L48 (R1b1a1a2a1a1c2b) 95+/-22 Gen., Frisian ~5,300-4,100 years ago
- R-Z9 (S268) R1b1a1a2a1a1c2b2 ~5,300-4,100 years ago
- R-Z30 (S271) R1b1a1a2a1a1c2b2a ~5,300-3,700 years ago
- R-Z32 R1b1a1a2a1a1c2b2a1
- Z2/S511, Z27 R1b1a1a2a1a1c2b2a1b ~4,600-3,500 years ago
- (R-S15510) R1b1a1a2a1a1c2b2a1b5, FGC23425/Y7415, S23231, Y7374
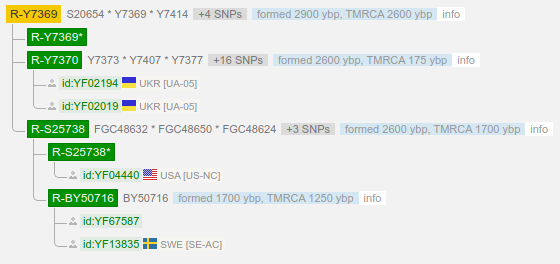
- (R-S8958) R1b1a1b1a1a1c2b2a1b5a Y7369 ~2,900-2,600 years ago
- (R-S15510) R1b1a1a2a1a1c2b2a1b5, FGC23425/Y7415, S23231, Y7374
- Z2/S511, Z27 R1b1a1a2a1a1c2b2a1b ~4,600-3,500 years ago
- R-Z32 R1b1a1a2a1a1c2b2a1
- R-Z30 (S271) R1b1a1a2a1a1c2b2a ~5,300-3,700 years ago
- R-Z9 (S268) R1b1a1a2a1a1c2b2 ~5,300-4,100 years ago
- R-L48 (R1b1a1a2a1a1c2b) 95+/-22 Gen., Frisian ~5,300-4,100 years ago
- R-U106/S21 (R1b1a1a1a2a1a1), ~5,400-4,500 years ago
- R-M269 (R1b1a1a2)
- R1b (R-M343)...
- R1 (R-M173), Central Asia, 12,000-25,000 years ago
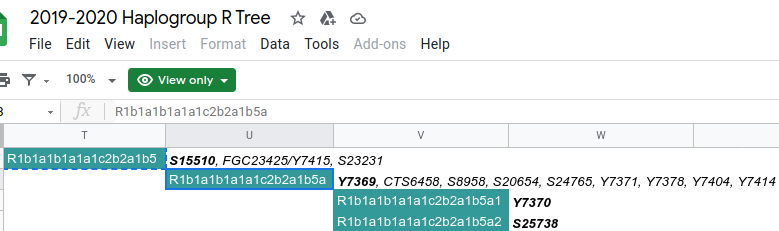
(Note that we may have further mutations but this the extent checked so far, as of 23 April 2015. This science is progressing fairly quickly and only recently was the R-L48 and the R-Z2 established as haplogroups. Tests taken: U106+, Z2+, Z31-, Z8-, ZU198-, L1-, P107-, P312-. In the diagram above there are intervening mutations but not of significant research interest. Currently we are terminal Z2 though there are some side mutations that have not been lined up to indicate whether future tests would be informative.
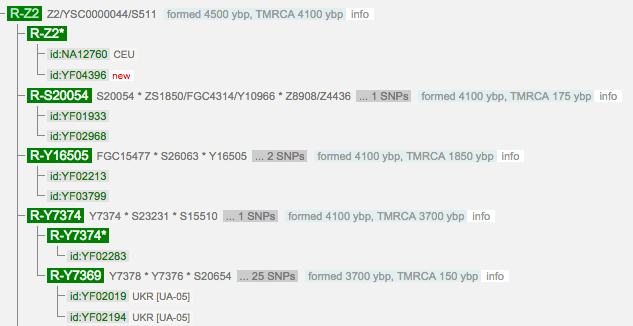
See additional experimental subclades of Z2 (excluding Z7) as of 29 Oct 2015:
Compare this with the developments that have occurred by 06 Jun 2016, less than 9 months later:
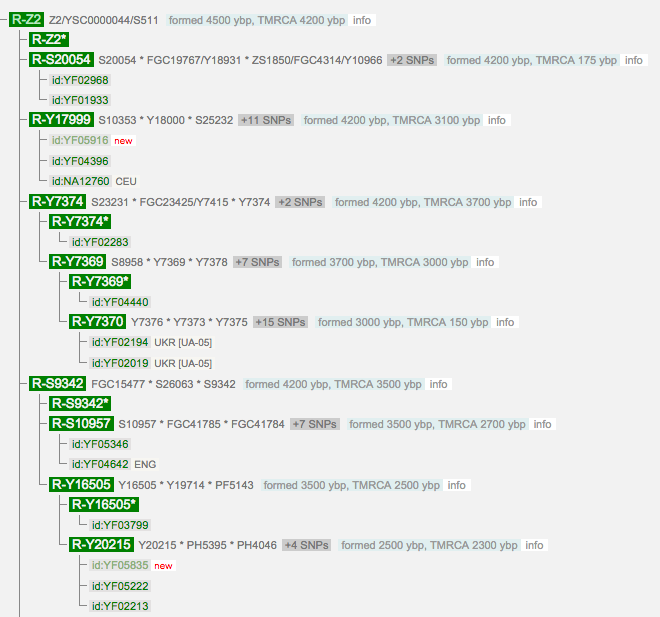
As of 06 Jun 2016 there are tests at YSEQ for S9342 and S20054 (but not Y17999 or S9342). YSEQ is the most advanced lab, the most up-to-date analyses available, and has the best analysis prices ($ 17.50 USD per SNP).
YSEQ also has a Wish a Panel custom panel, choosing from at least 7 existing SNP or STR markers. For example, once enough SNPs are available under Z2, it would be great to have a panel that could sort people easily. Then all the Z2 folks at FTDNA could head over and get an affordable advanced SNP sort. This could speed up discovery.
Note 08 Aug 2016 - One of my close genetic relations had a terminal SNP (which may or may not stay that way) identified as S15510, which is exciting as it is below Z2, but parallel with Z7, just where we should be looking.
If I read this correctly, and the entry has been added to the main R Haplogroup Tree, we are now officially (so far):
> (R-S15510) R1b1a1a2a1a1c2b2a1b5, FGC23425/Y7415, S23231
There is currently another haplogroup beneath this one, and two haplogroups beneath that. We could be terminal to the S15510 or in any of the three others currently known, or in any given unknown haplogroup beneath our current one.
Note: Y-SEQ puts the haplogroup for S15510 at R1b1a1a2a1a1c2b2a1 though the b5 has been assigned subsequently (though may be revised in the future).

Note 03 Sep 2016 - I've got confirmation on the S15510 test via Y-SEQ. I've inquired if there are additional downstream tests that are recommended. I've also upgraded my STRs from 69 to 111 at FTDNA, which can help with increasing the accuracy of MRCA for some of my matches that also have results for 111 markers. Note that S15510 is named R-Y7374 in YFULL YTree.
Note 30 Jun 2017 - We've gotten back a negative confirmation on the S20654 test (aka R-Y7369), which means we are not under that mutation, though there is still work being done for downstream mutations under S15510. In the yfull tree, this mutation sits below another intermediate, namely R-Y7378, which also has a sibling R-Y7374.
Note 2018 - Positive confirmation on Y7369 / R-S8958. This is now separate from the S20654 haplogroup either before this latest test or during ongoing research. I have a negative for S20654 but positive for Y7374. A bit confusing, but it is now clear that this is now the deepest (root) Haplogroup I have tested positive for, though there are more emerging haplogroups underneath. Step by step we get there.

Therefore officially:
> (R-S8958) R1b1a1b1a1a1c2b2a1b5a Y7369
Note 16-Sep-2019 - There are two more clarifications on related people, with (at least currently determined) terminal SNPs of BY41566 (grouped with BY41953, BY177933, BY145955, BY42115, BY41834, BY106970, BY41634, BY42105) and BY74109 (grouped with BY131220). The latter is a subclade of the former. These SNPs are situated three places below the S15510 SNP. In the future these groupings may be teased apart or reordered, but for now that is what the current status is. I'm leaning toward dong a BY74109 test. If positive, this would place me at the deepest known SNP, and if negative, that would also be informative as to what to test next (namely BY41566). However, none of these tests (individually) are available, currently. It appears that FTDNA has an improved lower level test of S8958 which is a step or two below the Y7378 available at Y-SEQ. Note that the YFull YTree has not caught up or harmonized with the FTDNA work. The FTDNA SNP prices are $39 each while Y-SEQ is $18, though I'm getting all my match data and insight from FTDNA.
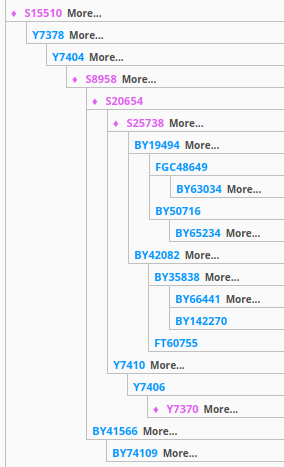
Note also that Y7369 has a much more recent appraisal than previously provided, and is now thought to be as recent as ~2,900-2,600 years ago.
Frisii, Angeln, and Saxon
Note that in general, we are dealing with disjointed timescales. That is, the mutation ranges (which are themselves quite large) are not in written history for the locations of these peoples. That is, the time of the mutations do not align with historically recorded events. However, the location of the mutations -- where they existed after taking place -- can be tracked to recorded history.
S21/U106 is considered the proto-germanic branch of the Indo-European family tree. However, not truly germanic in location, it is rather centered more north-central-western orientation. The maximum presence found around the Netherlands along with a so-called Frisian branch. "The Frisians and Anglo-Saxons disseminated this genetic mutation to England and the Scottish Lowlands."2
The Frisians, a germanic tribe, are first mentioned by Roman writers around 2,000 years ago. The Angeln (Angles) and the Saxons were also both Germanic tribes, and whose source of information is also Roman writers, specifically Tacitus' Germania originally composed in 98 CE.
The people known as The Frisii or Frisians migrated to Frisia, modern day northern Netherlands, around 2,500 years ago. While identified by our L48 haplogroup markers, the exact date of this mutation could have been before or after. However, the location of Frisia is indeed the center of the prevalence of the mutation. This is more or less one of the homelands our ancestors had.
Modern day Friesland is a province in the Northern Netherlands. Friesland is largely agricultural, with Frisian cattle and horses, as well as sea and island tourism. There are nearly 200 windmills in Friesland.
The Anglo-Saxon mass migration occurred roughly 50 generations ago (1,500 years at 30 years/generation). This migration can be traced by the L48 mutation that we possess (along with others we don't, e.g., L47, L21).3
See also brief articles from the History Files:
Haplotype Groups and MRCA Relations
The methods for non-specialist use of genetic genealogy is twofold: In terms of haplogroups, we can determine what larger genetic group we belong to and place that geographically (and somewhat historically). In this case, for the McNeill, we can see our L48 mutation as being a part of the Angles/Saxons/Frisians group. Subsequent historical knowledge of this group gives some indication of how and when (in rough terms) our genetic ancestors got to where we are today.
Secondly, by comparing most recent common ancestors (MRCA) with other individuals, we can see where our ancestors and their relations have lived (subsequent to the larger group migrations).
Combined with genealogy, we can compare genetics with names, and also do research with standard genealogical tools, such as birth, marriage, and death certificates, headstones, property deeds, lawsuits, and the like.
Genealogically we can get back to our immigrant ancestor. Genetically (doing comparisons between tested individuals) we can see some evidence for Ulster (and lack of evidence with island and highland Scots, as well as other groups such as the Irish). Genetic Haplogroup testing provides us with larger group membership.
Note that this grouping does not exclude the possibilities such as having Danish Viking blood (since the Danes were indistinguishable from the Frisians, Angles and Saxons by the latter's migration to southern Denmark). Also in all of this the female line is not considered, which is the other half of the story (yet to be told).
The Angles and the Saxons, from which we have the term Anglo-Saxon, as well as Frisians are believed to have been located in present-day Southern Denmark/Northern Germany (precisely Schleswig-Holstein) prior to their migration to Briton.
The Frisians and the Anglo-Saxons had similar languages and political organizations. Old Frisian is the most closely related language to Old English. Frisians, Angles and Saxons are genetically indistinguishable by current findings.
Jumping ahead of a few mutations our ancestors inherited the Z2 mutation (R1b1a1a2a1a1c2b2a1), and we may have further, more recent haplotype mutations, as the science is still ongoing.
Power and Progeny
There are two specific lessons to learn about the diffusion of Y-DNA, one is the use of power by a single individual, and his progeny, to create a horde of descendants. This is most illustrated by great warrior king Niall of the Nine Hostages. Niall Noígíallach was an Irish king who lived around 1600 years ago (about 400 CE). His specific Y-DNA is estimated to be present in about 3 million living (male) persons, including up to 21% of Northern Ireland and 2% of the state of New York.
Even with that impressive count, Niall holds second place in history. Genghis Khan beats him out with approximately 16 million living heirs. To reach this level of propagation wholesale slaughter of his opponents and rapine and a harem culture, along with a multi-generational power structure which provided ongoing propagation.
Genghis Khan beats Niall Noígíallach because Genghis Kahn conquered at a much later date (approximately 800 years), had greater technology and wider ambitions. But what about our own more modest ancestors? They did not conquer in terms of vast lands, but they did produce viable progeny, and their actions are why we are here today.
In order for these two warlords to produce such progeny they needed exclusive access to a significant amount of fertile females. The converse is that they needed to deny access to other fertile males. There can be two approaches to this. The first is killing large numbers of males. The second is a culture which enforces the same kind of access. With Genghis Kahn, the vast areas and populations he conquered (along with the killing of the men), along with a dynasty which lasted many generations, ensured procreative effectiveness.
For Niall of the Nine Hostages, we have much less to go on, but the same kind of conditions likely held. Kill and subjugate the male competition and reinforce for generations a culture which provides the ruling kings and their family with ongoing and widespread access to fertile females.
The cultural aspect has been called a Sexual Apartheid by Mark Thomas of University College London.4 He has applied this term to the whole of Anglo-Saxon Britain, where in the settled towns a complete domination of the migrating groups' Y-DNA in the population. He claims that to make sense of the current genetic evidence, there was historically:
> a sexually biased, ethnically driven reproductive pattern, in which Anglo-Saxon males fathered children with Anglo-Saxon females and possibly Celtic females, while the reproductive activities of Romano-Celtic males were more restricted, is the most plausible explanation for the demographic, archaeological, and genetic patterns seen today.
Most recently, the degree of genetic domination is considered important but less severe than Thomas suggests.
Genetic Genealogy and Migration
If all genetic markers were to anchor a people in a place then interpretation of genetic genealogy would be identical to reading the earliest histories of a people. Of course things are not so straightforward. An excellent example is the genetic genealogy of the variety of people who have lived in the Xinjiang Autonomous Region of China. Well-preserved mummies from up to 4,000 years ago have revealed a story that has political implications in China's occupation of Xinjiang. The government propaganda posits Han chinese settlement and occupation which is disputed by a variety of archealogical evidence, including dna analysis which has been suppressed in the Tarim Basin.
Some of the Tarim Basin people were a celtic people who wore the oldest known tartan textile.
> The oldest tartan ever found was worn by the Ürümchi mummies around 1500 BC. The Ürümchi mummies belong to a group of caucasian indoeuropeans who explored the Silk Route between Europe and Central Asia. The mummies were found in the Xinjiang region, western China, dressed with colourful tartan plaids.
McNeill and Variants
Genetically Anglo-Saxon, there is a question of how did we get the McNeill name. There is an interesting look at distribution of the name and its spelling variants. There are several locations that match up with Angeln presence in Bernecia / Northumberland (East Lothian) which came under Scottish rule, as well as Ayrshire and Argyll and most tellingly Northern Ireland. This matches up with a potential migration over generations.
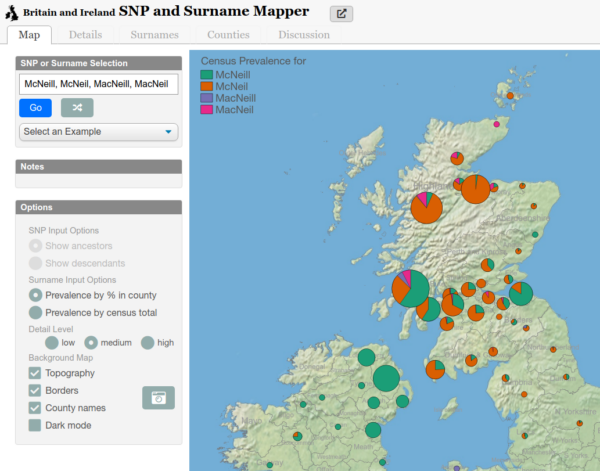
Note the spelling variants as well.
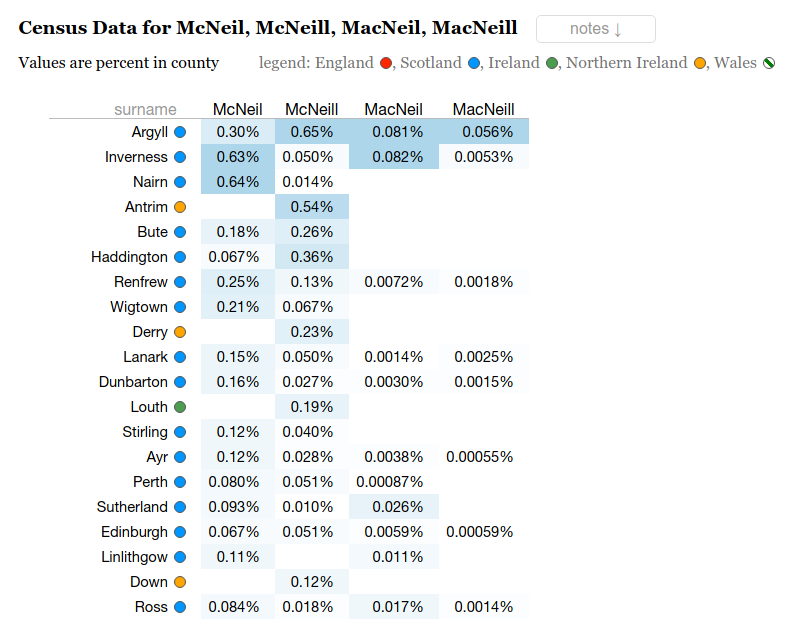
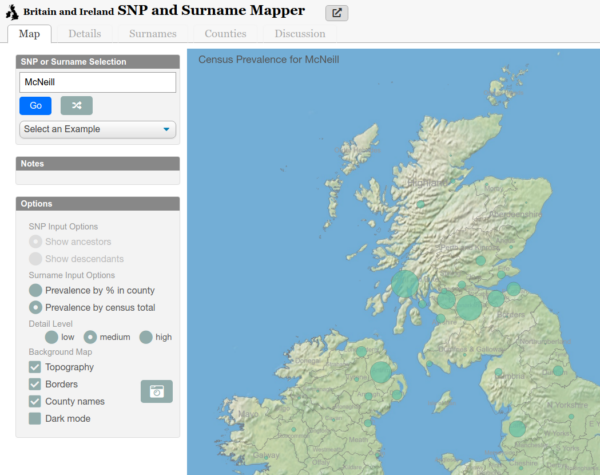
A vague trajectory over time...

Back to the table of contents for McNeill and Beyond - A Memoir